Biofilm model systems
The LMM develops and undertakes studies investigating the mechanisms of biofilm formation for all forms of bacteria.
We are currently defining patterns of gene expression in planktonic, biofilm, and persister cells, confirming the role of candidate genes essential for biofilm formation by genetic screening of random insertional inactivation libraries, and validating promising targets by inactivation and complementation strategies.
Our research line on biofilms and bacterial pathogenesis has generated great interest from the pharmaceutical industry, resulting in several service and research collaborations.
Static Assays
Static assays are high-throughput systems that are characterised by limited nutrients and aeration but enable direct rapid quantification of biofilm mass.
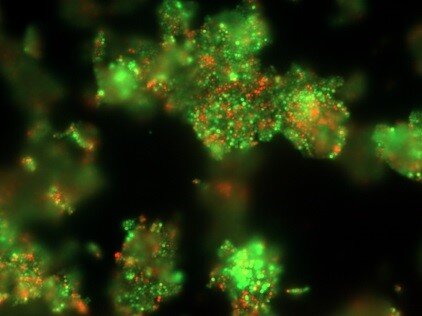
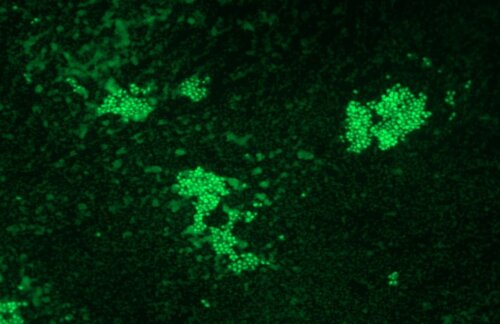
PEG-plate biofilm system: bacterial cells are grown on a coverlid that is composed of pegs that fit into the wells of the microtiter plate containing the growth medium and bacteria. Staining will give information on the total biofilm mass.
Dynamic Assays
Dynamic assays are medium throughput systems where spent culture is constantly replaced by fresh medium, allowing for the control of environmental parameters.
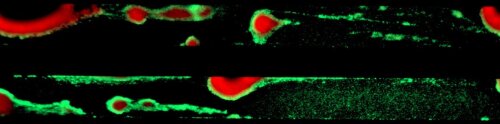
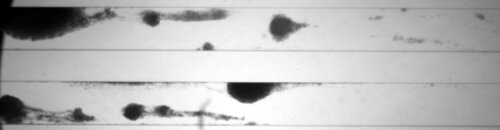
Bioflux system: biofilms are grown using a device comprised of microfluidic channels and a distributed pneumatic pump that provides fluid flow. Biofilm formation can be followed and quantified by light microscopy or a wide range of fluorescence microscopy stains.