The molecular and phenotypic basis of rapid adaptation to heavy fishing

Besides their role as a model system in speciation research, Lake Malawi cichlids are food for millions of people, and subject to a recent and strong increase in fishing. Some cichlid populations have seemingly adapted their life histories to high fishing pressure and mature at substantially smaller size compared to less heavily fished populations. In this project, we will dissect the genetic factors contributing to rapid phenotypic adaptation in Lake Malawi cichlid fish populations following ~40 years of extremely heavy fishing.
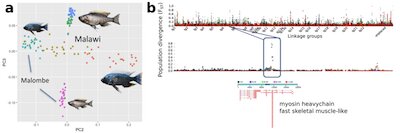
Fisheries-induced adaptation (FIA) in Lake Malawi cichlids provides an ideal system to study rapid adaptation, because of
- extensive genomic resources currently becoming available for Lake Malawi cichlids will allow us to cost-effectively sequence and compare the genomes of weakly and heavily fished populations
- > 2000 already existing Malawi cichlid whole-genome sequences of 270 species will allow us to infer the long-term evolutionary history of genetic variants used in recent adaptation to heavy fishing
- analysing population genome data from museum specimen from before and during heavy fishing, will give unprecedented insight into the selective process across time
breeding cichlids in the lab will allow us to
- experimentally quantify genetic (as opposed to environmental) differences in traits implicated in fisheries-induced adaptation
- map the genetic variants contributing to within and between-population variation in traits using association mapping and crosses.
In summary, the combination of genome sequencing of recent and historic natural populations with controlled breeding experiments will greatly advance our understanding of the link between selection pressures, phenotypes and genotypes, will enable the dissection of complex traits, and will uncover how genomes can rapidly adapt to human-induced selection.
The origin of old genomic variation and its role in recent adaptation
We have recently found evidence that Lake Malawi cichlids harbour genomic regions of exceptionally high genetic diversity (Svardal et al., in preparation). We are now investigating the origin of genetic variants contributing to this diversity and their role in adaptation and diversification.
Furthermore, we recently found that two divergent lineages hybridised in the ancestral population of Lake Malawi cichlids. Was this hybridisation event the source of high diversity regions, or are high diversity regions rather produced and maintained by balancing selection? Are variants in these genomic regions used in recent adaptation?
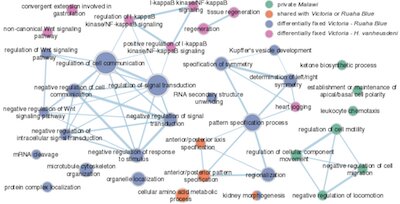
The gene ontology network shows biological processes enriched in genes with genetic variation derived from an old hybridisation event (in blue).
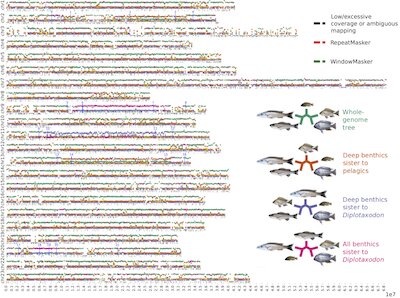
Genomic patterns of cross-species genetic exchange and its role in ecological adaptation
Is hybridisation between species and exchange of genes a rare or common process and how important is it for adaptation? Recent evidence from all kinds of organisms suggests that gene flow between species is common. We aim to quantify the genomic patterns of genetic exchange between cichlid fish species and infer its role and function in adaptation and diversification.
The domestication history of Siamese fighting fish and the genetic basis of aggression
Siamese fighting fish (Betta splendens) are popular ornamental fish that have recently been selectively bred for different fin shapes and colours. However, in South East Asia they have already been domesticated for 100s of years for show fights, with the result that domesticated strains are much more aggressive than their wild counterparts. In collaboration with Andrés Bendesky (Columbia University), Richard Durbin (University of Cambridge), and Lukas Rüber (Natural History Museum, Berne) we are analysing genome and transcriptome data to understand the history of domestication of fighting fish and the molecular basis of their aggression.
